I was recently talking with Geof Hannigan, a postdoc in our lab, and he pointed out a pretty critical piece of information I had been overlooking. When sequencing reads become long enough, you may sequence passed the fragment size and into the 3’ adapter. This is especially problematic when you move on to assembly because these introduced sequences will have 100% homology to one another and assemble on the adapter instead of actual genome sequence. Similar problems can arise in a couple different ways as shown in the figure below. These artifacts will leave you with contigs that don’t reflect biology at all.
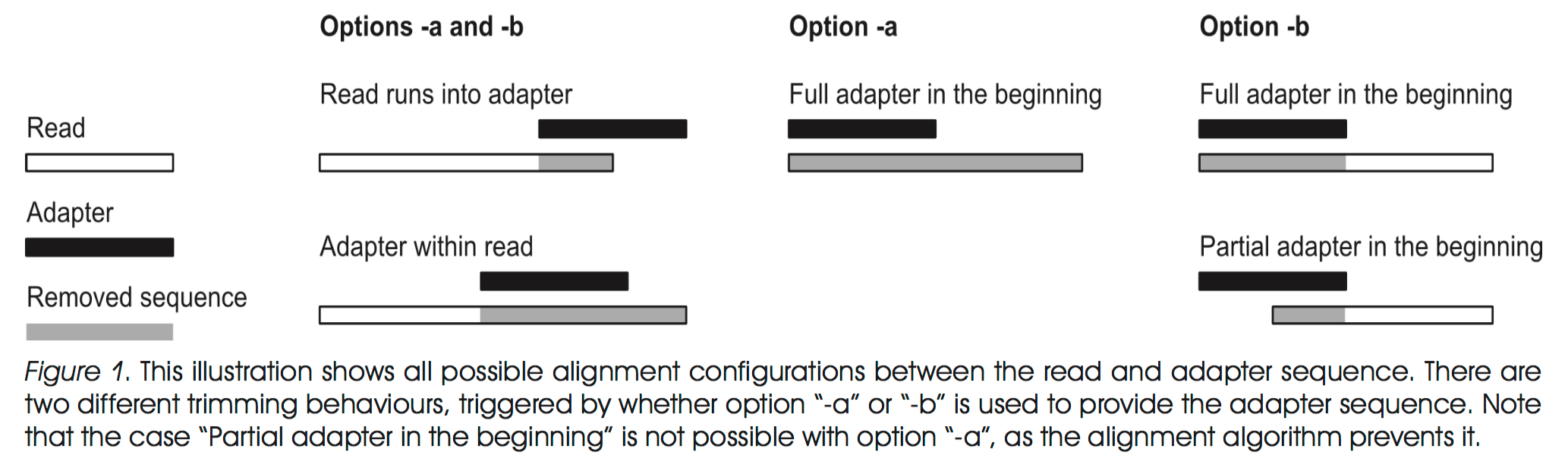
To avoid this I am now using a program called Cutadapt, where the figure came from, to remove any residual adapter sequences from my reads prior to assembly. The program requires python 2.7 and the command to run it looks like this:
1
python2.7 /mnt/EXT/Schloss-data/bin/cutadapt-1.9.1/bin/cutadapt --error-rate=0.1 --overlap=10 -a CAAGCAGAAGACGGCATACGAGATTAAGGCGAGTCTCGTGGGCTCGG -A GACGCTGCCGACGAGCGATCTAGTGTAGATCTCGGTGGTCGCCGTATCATT -o read_1.trimmed.fastq -p read_2.trimmed.fastq read_1.fastq read_2.fastq
Each argument supplies the following info:
1
2
3
4
5
6
7
8
--error-rate = Rate for how generously you still call a matching sequence, basically 1 in 10 bases can be a mismatch
--overlap = Minimum number of overlapping bases to call a match
-a = 5' sequencing primer full sequence, adapter + index
-A = reverse complement of the 3' sequencing primer
-o = Output file name for read 1 followinf trimming
-p = Output file name for read 2
The last two file names without options before them are the forward and reverse read files
In order to get the reverse complement of the 3’ adapter, I wrote a small python script to reverse the order of a nucleotide sequence and then switch each base to it’s complement. Below is what it looks like and I’ve hosted on the Github page I made for scripts I talk about on my blog. This new adapter trimming step is critical for both my metagenomic and metatranscriptomic pipelines, basically anything with some amount of random fragment size. Otherwise the primer fragments left in the data negatively impact and assembly or mapping you try to do downstream.
#!/usr/bin/env python
# Computes the reverse complement of a given nucleotide string
# USAGE: rev_comp.py input_sequence
import sys
def reverse_complement(nucleotides):
seq_str = str(nucleotides).upper()
rev_seq_str = seq_str[::-1]
pyr_seq_str = rev_seq_str.replace('A', 'n')
pyr_seq_str = pyr_seq_str.replace('T', 'A')
pyr_seq_str = pyr_seq_str.replace('n', 'T')
final_seq_str = pyr_seq_str.replace('C', 'n')
final_seq_str = final_seq_str.replace('G', 'C')
final_seq_str = final_seq_str.replace('n', 'G')
return final_seq_str
print(reverse_complement(sys.argv[1]))Below are some stats from the assembly of the same 250 bp paired-end dataset from a HiSeq, just with and without the trimming protocol I laid out above.
Without 3’-adapter trimming:
1
2
3
4
5
6
7
8
9
10
11
Total sequences: 431204
Total bases: 495.00 Mb
Sequence N50: 1762
Sequence L50: 28
Sequence N90: 452
Median sequence length: 560
Interquartile range: 436
Shortest sequence length: 250
Longest sequence length: 420252
Sequences > 1 kb: 88470
Sequences > 5 kb: 10866
With adapter trimming:
1
2
3
4
5
6
7
8
9
10
11
Total sequences: 496917
Total bases: 471.39 Mb
Sequence N50: 1434
Sequence L50: 44
Sequence N90: 369
Median sequence length: 474
Interquartile range: 423
Shortest sequence length: 250
Longest sequence length: 201752
Sequences > 1 kb: 83535
Sequences > 5 kb: 10603
The assembly quality doesn’t seem to have changed dramatically, however the best metric is really how many reads map to the contigs that you just assembled. Prior to this trimming <49% of reads mapped at least once to a contig. Now >70% are mapping! That’s a huge improvement! The new contigs should reflect what I actually sequenced and allow for a better analysis from now on.