Our paper on modeling context-specific metabolism of C. difficile is finally accepted in mSystems! In it we generated and extensively curated a C. difficile GENRE for a hypervirulent isolate (str. R20291). In silico validation revealed high degrees of agreement with experimental gene essentiality and carbon source utilization data sets. Then in collaboration with Dr. Rita Tamayo at UNC, whose research focuses on an evolutionary strategy in some bacteria for phenotypic heterogeneity and virulence known as phase variation. We utilized transcriptome sequencing from distinct phases of C. difficile, and in combination with our previously published integration algorithm, we created context-specific models of metabolism to identify possible metabolite signals that drive differences in virulence.
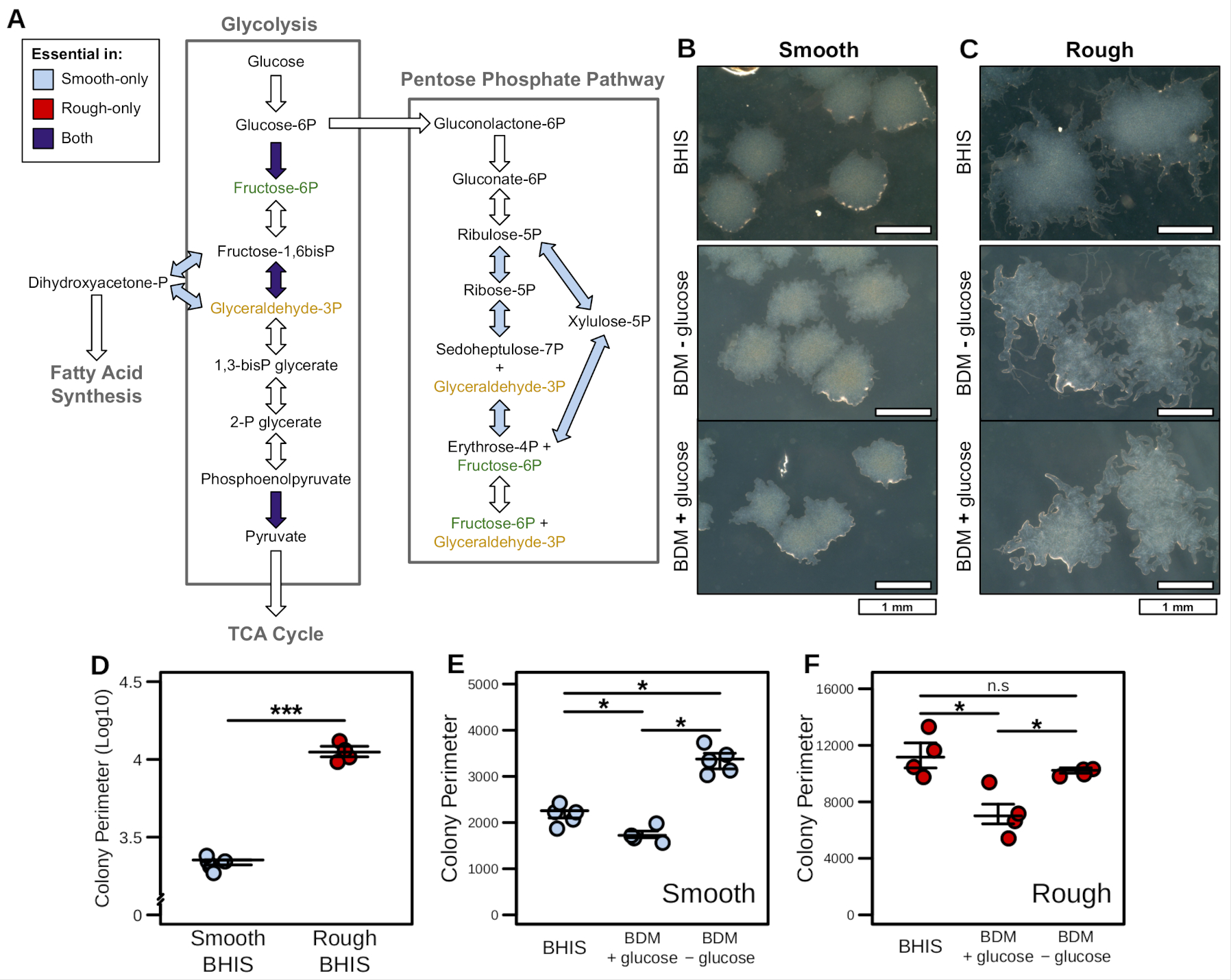
Using this GENRE-based analyses we found that glucose utilization through the pentose phosphate pathway is essential in the smooth phase variants of str. R20291. Here we show (A) Gene and reaction essentiality results for glycolysis and the pentose phosphate pathway across both the rough and smooth phase variant context-specific models. Components were deemed essential if models failed to generate <1% of optimal biomass flux. (B and C) Colony morphologies resulting from smooth and rough variants of C. difficile str. R20291 grown on either BHIS or BDM ± glucose after 48hr of growth. Defined medium colonies were then subcultured onto BHIS medium for an additional 24hr as indicated. Increased colony perimeter was found to be the defining characteristic of the rough colony morphology. This feature was quantified for multiple colonies under each permutation of colony variant and growth medium. (D) Colony perimeter for smooth and rough progenitor colony variants grown on BHIS. (E and F) Smooth (E) or rough (F) colony variant perimeter during subculture onto each of the BDM carbon source medium formulations.
Our results support that differential C. difficile virulence is associated with distinct metabolic programs related to use of carbon sources and provide a platform for identification of novel therapeutic targets.