After a long review process, the final publication from my dissertation work is finally published! In it were able to show that not only did Clostridium difficile differentially alter the transcriptional programs of communities it was invading, but that the degree of change positively correlates lasting infection. What this means is that those individuals where C. difficile infection is more persistant may possess a microbiota that is more susceptible to changes induced by the pathogen. We accomplished this by comparing the metatranscriptomes of mock and infection mouse intestinal communities across multiple antibiotic pretreatments that each result in equal initial susceptibility to infection but differ in resultant community structure and rate of pathogen clearance. You can see one example of changes caused by C. difficile to streptomycin-pretreated gut communities below.
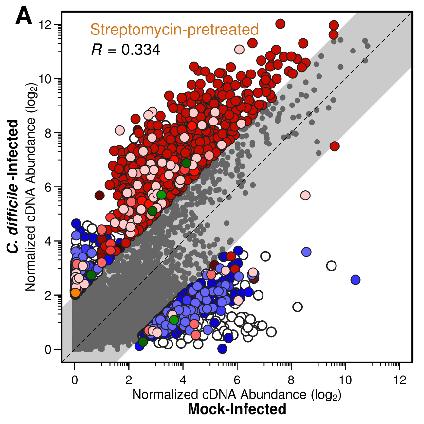
This figure illustrates metatranscriptomic expression of many genes in the community are more highly upregulated in the absence of C. difficile and that genes primarily belong to bacteria within the genus Lactobacillus. What we found interesting about this finding was that streptomycin results in a gut community where Lactobacillus represents less than 1% of the detectable bacteria. This proved to be the case across each antibiotic pretreatment tested, where the minority members of the community appear to be more greatly impacted by infection than those taxa that dominate those communities. This was quantified below by combined the metatranscriptomic results with 16S rRNA gene amplicon sequencing relative abundance data.
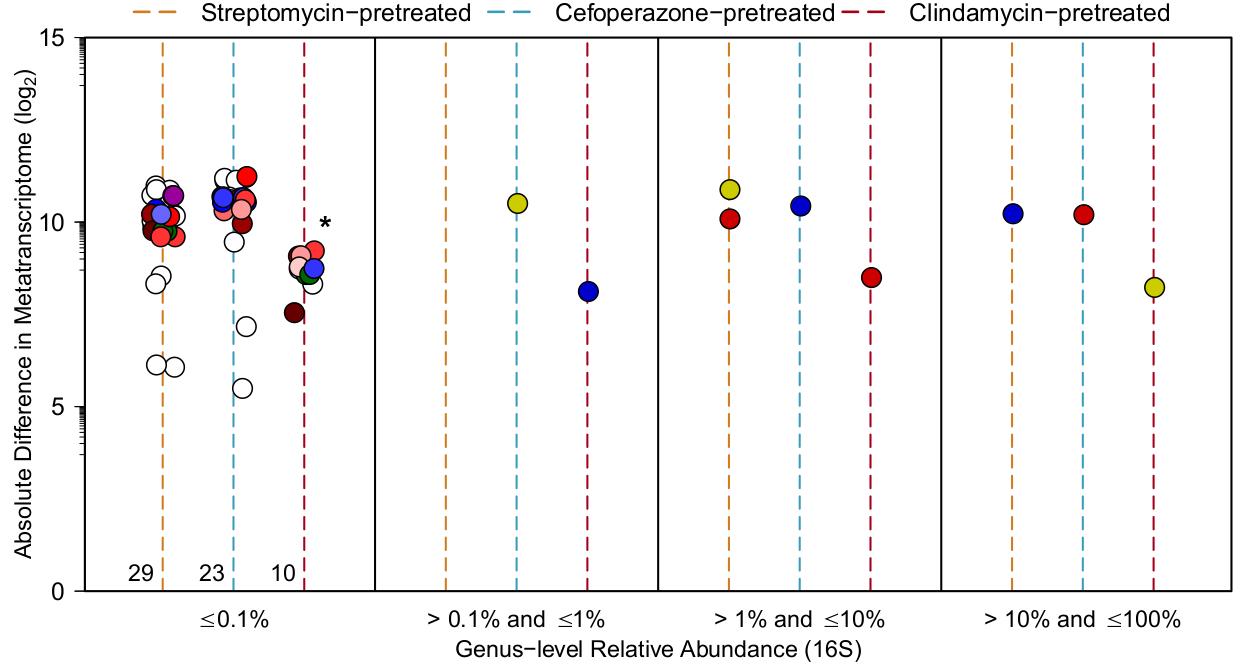
What this shows is that those taxonomic groups of bacteria that are more greater affected by C. difficile infection seem to be always in underrepresented in each community. You could say that C. difficile is attacking the loser is each of these instances to try and promote it’s own existence, a pretty low blow even for a pathogen. There are instances similar to this in bacterial communities, but this is the first time it has been demonstrated in the context of a bacterial infection in a host-associated community of microbes.